 Scientists report the genomic landscape of the Mediterranean basin and the Greater Middle East, a central hub for ancient civilizations and home to approximately 10 percent of the World population, in an article published in the September 2016 issue of Nature Genetics. Project results provide important insights into two major areas of human genetics, namely the identification of disease-associated genes and the elucidation of the human migration pathways from Africa to Asia and Europe. The high consanguinity rates, a characteristic of Middle Eastern populations, enhance disease gene identification by 4 to 7 fold.
Scientists report the genomic landscape of the Mediterranean basin and the Greater Middle East, a central hub for ancient civilizations and home to approximately 10 percent of the World population, in an article published in the September 2016 issue of Nature Genetics. Project results provide important insights into two major areas of human genetics, namely the identification of disease-associated genes and the elucidation of the human migration pathways from Africa to Asia and Europe. The high consanguinity rates, a characteristic of Middle Eastern populations, enhance disease gene identification by 4 to 7 fold.
The Project was first proposed by Prof. Dr. Tayfun Özçelik from the Department of Molecular Biology and Genetics and an international group of scientists who convened at Bilkent University in 2009 and 2010. The project was conducted over a period of 5 years and with a budget of tens of millions of dollars. More than 180 scientists from Algeria, Egypt, France, Israel, Iran, Iraq, Jordan, Lebanon, Libya, Morocco, Pakistan, Qatar, Saudi Arabia, Syria, Tunisia, Turkey, United Arab Emirates and United States contributed to the project. Rockefeller, Harvard, MIT, California, Cornell, INSERM, Alfaisal and İstanbul Universities were amongst the partners. The honor to present the results of project to the scientific world was given to Prof. Özçelik through a “news and views” article5 also published in the same issue of Nature Genetics.
As a first step, scientist, including 30 from Turkey, formed the “Greater Middle East Variome Consortium” and established a DNA bank of 1,111 individuals whose genomes were sequenced.
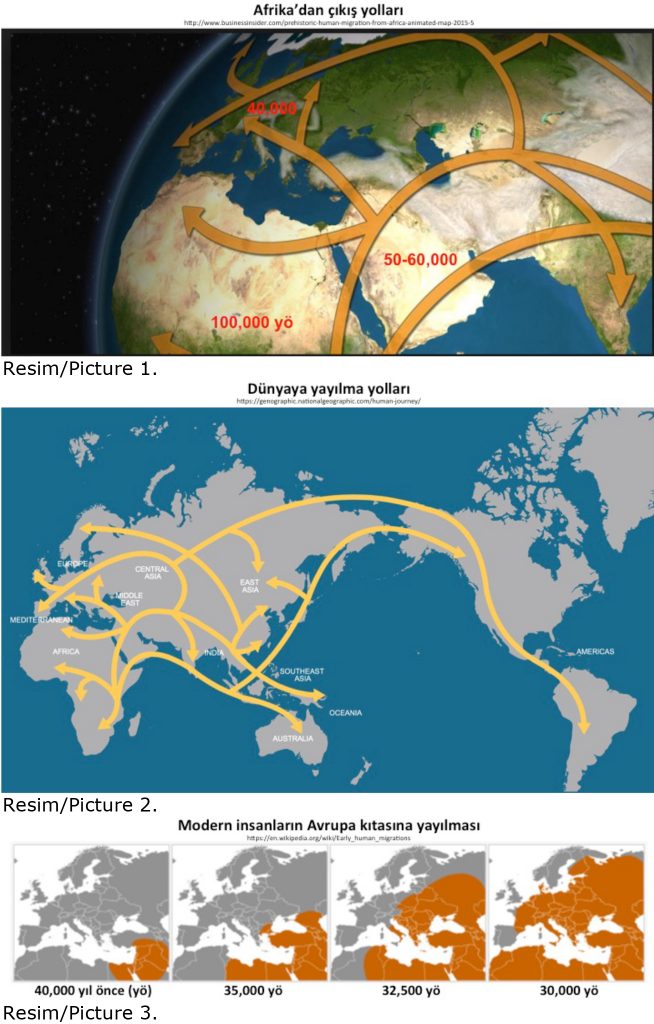
Migration pathways and human history
Scientists first compared the genetic structure of Middle Eastern populations with that of “1000 Genomes” populations from Europe, Asia, Africa, Australia and America. They found significant differences amongst the Middle Eastern populations, which are spread over a large geographic region from Northwest Africa to Southwest Asia. They discovered clusters with Asian or European populations. Patterns of human migration and drift were recapitulated, which follows a route from Africa to Mesopotamia and Anatolia, to Arabian Peninsula, Iran & Pakistan to India and through oceanic islands all the way to Australia. This journey covers a period of over 100 thousand years (Pictures 1 & 2).
Concurrently, important insights into the genetic diversity of Anatolian populations were documented including a flow of humans from Anatolia to Europe. Historic evidence points to the central importance of Göbeklitepe and present day Urfa, Mardin, Gaziantep regions in the spread of ancient civilizations. While the inter genetic distances between Northwest Africa and Arabian Peninsula or Iran and Pakistan were found to be significant just like the differences between Finnish (Finland) and Toscani (Italy), highest levels of European admixtures were observed for Turkish peninsula followed by Syrian Desert (Picture 3).
Identification of the genetic bases of inherited diseases
Genome Projects have attained great scientific importance during the past two decades. From this point onwards, identification of disease associated genes, development of diagnostic tests and targeted treatment modalities have become the foundation stones of precision medicine initiatives. Precisely at this point, the family structure of populations becomes all the more relevant to understand the genomic landscape of humans. More specifically, large families with many children and consanguinity amongst the parents in the extended kindred facilitate dissection of normal genomic diversity from disease causing mutations. For example, homozygosity blocks over 4 megabases, a hallmark of disease associated genetic changes, are observed exclusively in the Anatolian, Mesopotamian and Middle Eastern populations. This is regarded as a unique opportunity to fully exploit the potential of advanced and technology-driven genome interrogations to mitigate the burden of genetic diseases and serve humanity. More precisely, families from Anatolia facilitate the identification of faulty genes by 4 to 7 fold. This means it becomes 700 percent more plausible to identify the culprits in the Anatolian Peninsula when compared to America, Europe or China.
A new hypothesis
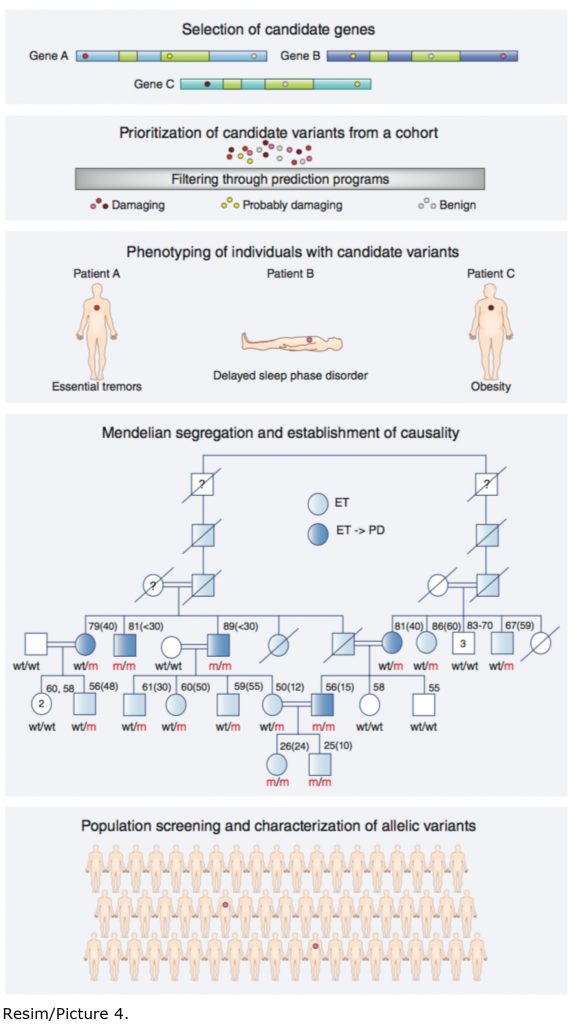 It is a well established fact that genomics has been spectacularly successful during the past 20 year when it comes to the identification of rare diseases such as cystic fibrosis, phenylketonuria, ataxia or inborn errors of metabolism. While rare diseases are individually rare as the name implies, they together constitute the largest group of disorders -now totaling more than 5000- that affect children in the developed world. However, when it comes to the adults, rare diseases subside and complex diseases such as obesity, hypertension, diabetes, migrane, sleep problems, neurodegeneration and certain types of cancers take over. But unfortunately the genetic mutations that ultimately lead to these complex diseases are not known by and large. In other words, success stories like the ones on rare diseases are yet to be written. Scientists recognize that the main bottleneck is at a conceptual level rather than technological constraints. In other words, it is necessary to develop new strategies to interpret the sequences that constitute a genome. In the Nature Genetics news and views article that Dr. Özçelik wrote together with his postdoctoral fellow Dr. Emre Onat, a new conceptual framework and an analysis path is presented to decipher the molecular bases of complex diseases. The duo propose that the strategy to identify the genes associated with Uner Tan syndrome first described in quadrupedal families from Turkey a decade ago, or more recently the gene that leads to essential tremors and Parkinson’s disease, all identified at Bilkent, could be extended to a plethora of complex phenotypes (Picture 4).
It is a well established fact that genomics has been spectacularly successful during the past 20 year when it comes to the identification of rare diseases such as cystic fibrosis, phenylketonuria, ataxia or inborn errors of metabolism. While rare diseases are individually rare as the name implies, they together constitute the largest group of disorders -now totaling more than 5000- that affect children in the developed world. However, when it comes to the adults, rare diseases subside and complex diseases such as obesity, hypertension, diabetes, migrane, sleep problems, neurodegeneration and certain types of cancers take over. But unfortunately the genetic mutations that ultimately lead to these complex diseases are not known by and large. In other words, success stories like the ones on rare diseases are yet to be written. Scientists recognize that the main bottleneck is at a conceptual level rather than technological constraints. In other words, it is necessary to develop new strategies to interpret the sequences that constitute a genome. In the Nature Genetics news and views article that Dr. Özçelik wrote together with his postdoctoral fellow Dr. Emre Onat, a new conceptual framework and an analysis path is presented to decipher the molecular bases of complex diseases. The duo propose that the strategy to identify the genes associated with Uner Tan syndrome first described in quadrupedal families from Turkey a decade ago, or more recently the gene that leads to essential tremors and Parkinson’s disease, all identified at Bilkent, could be extended to a plethora of complex phenotypes (Picture 4).